How To Register A 4d Image Fsl Diffusion
Preprocessing Diffusion Images
T1
T2
DWI
DWI vs. DTI
Diffusion weighted imaging
- Measures water move within a voxel of tissue
- Intensity value represents rate of water diffusion
Improvidence tensor imaging
- Specific type of modeling of diffusion weighted images based on the theory that water molecules diffuse differently within unlike types of tissue
Symlink
Preprocessing DWI creates a lot of files and anything that can be done to reduce the number of files is helpful. Create a derivative directory, pDWI, and instead of copying your files from BIDS, create a symlink.
cd ${DERIV_DIR}/${SUBJID}/ ln -s ${BIDS_DIR}/${SUBJID}/dwi/${SUBJID}_dwi.nii.gz dwi.nii.gz ln -southward ${BIDS_DIR}/${SUBJID}/dwi/${SUBJID}_dwi.bval dwi.bval ln -southward ${BIDS_DIR}/${SUBJID}/dwi/${SUBJID}_dwi.bvec dwi.bvec Verify Volumes
It is very piece of cake to have missing volumes in the DWI. It is critical that the numbers match between the NIFTI image and the bvec and bval files.
$ c4d dwi.nii.gz -info Image #one: dim = [104, 104, 69, 63]; bb = {[-103.445 91.427 -36.6199 0], [104.555 299.427 101.38 819]}; vox = [2, 2, 2, 13]; range = [0, 2910]; orient = Oblique, closest to RPI This NIFTI prototype has 63 volumes with a FOV of 104 x 104 ten 69. Make sure in that location's 63 entries in the bval file:
$ cat dwi.bval | wc -w 63 Finally, make certain you have the same number of volumes in bvec file:
$ head dwi.bvec | wc -w 63 Pad Paradigm
If your FOV box is right upwards against skull or even cutting off part of skull merely not brain, you will need to pad the image to give background space:
$ c4d \ dwi.nii.gz \ -pad 10% 10% 0 \ -o dwi_padded.nii.gz Mask B0
Must exist equally authentic as possible.
pnlNipype
Using nifti_bet_mask that packages FSL:
~/build/pnlNipype/exec/nifti_bet_mask \ -i dwi_bse.nii.gz \ --bvals dwi.bval \ -o dwi Using nifti_atlas that packages ANTs:
~/build/pnlNipype/exec/nifti_atlas \ -t dwi_bse.nii.gz \ --train ~/apps/pnlbwh/pnlNipype/trainingDataT2Masks-12a14d9/trainingDataT2Masks-hdr.csv \ -o dwi Make clean mask
Sometimes all that's needed is to erode / shrink the mask a little to make it better
c3d \ dwi_mask.nii.gz \ -erode 1 1x1x1vox \ -o dwi_mask_cleaned.nii.gz Eddy Current Correction
This correction volition help set up eddy electric current-induced distortions and subject movements during the scan.
Set acqparams.txt
All data can be derived / found in the DWI JSON file. For your acqparams.txt file, each row consists of a vector (three values) that specify what the phase-encode (PE) axis is. The fourth chemical element in each row is the time (in seconds) between reading the middle of the start echo and reading the center of the concluding repeat. The fourth element is also the reciprocal of the PE bandwidth/pixel. Remember that to calculate the reciprocoal you lot take: (1 BandwidthPerPixelPhaseEncode)
printf "0 1 0 0.095" > acqparams.txt 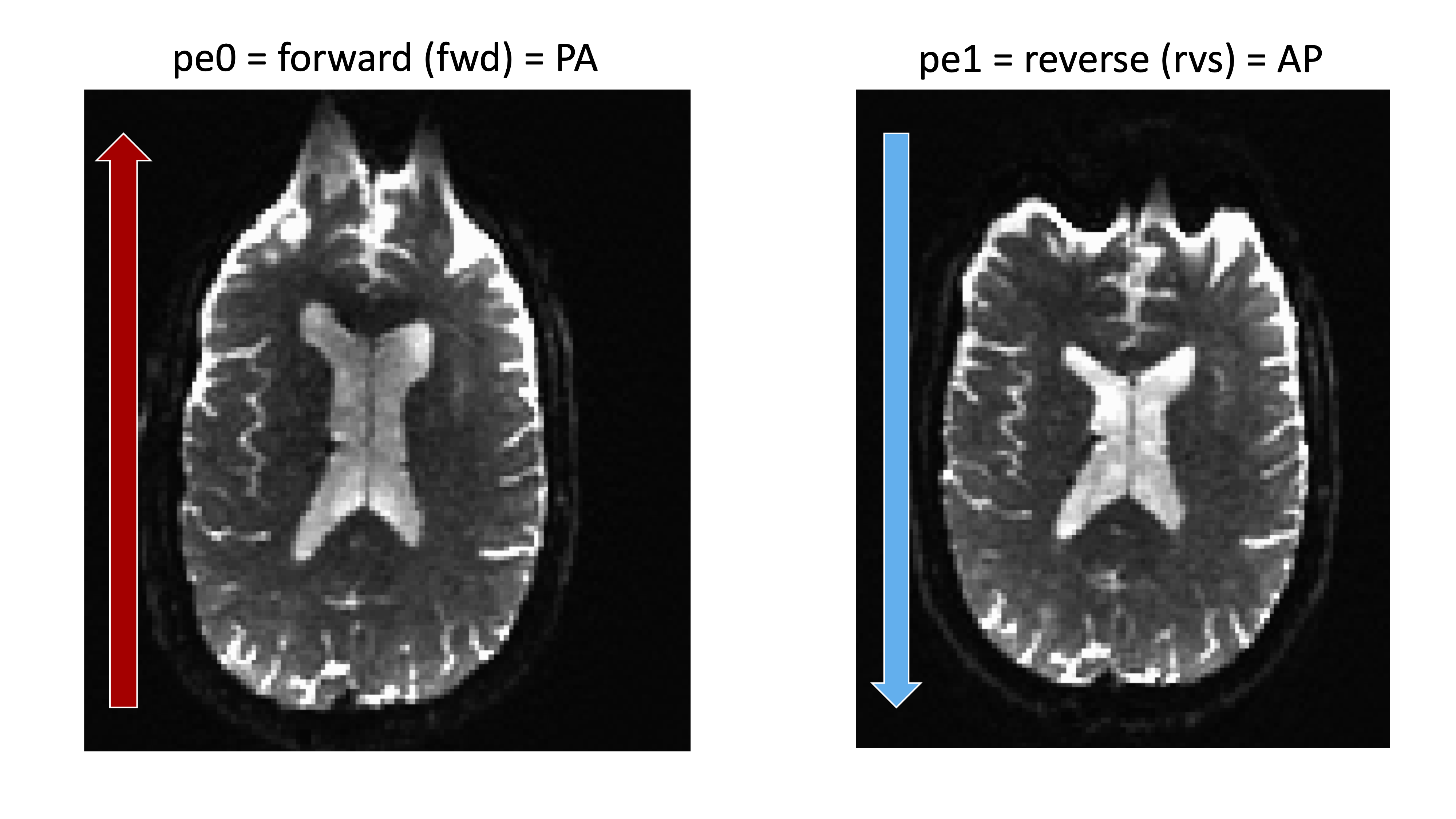
Forward
- Posterior to Inductive
- Phase Encoding is j
- acqparams.txt is 0 1 0
Contrary
- Anterior to Posterior
- Phase Encoding is j-
- acqparams.txt is 0 -1 0
Looking at the JSON file:
- "PhaseEncodingDirection": "j-,"
- "BandwidthPerPixelPhaseEncode": 13.935,
- (one = 0.07176175099)
printf "0 -ane 0 0.072" > acqparams.txt Fix index.txt
Create an index file that tells eddy which line of the "acqparams.txt" file to employ for each book of data. In our case above, the index file has a single line with 63 i's separated by spaces:
myVar = ($( wc -w dwi.bval)) indx = "" for (( i = 1 ; i <= $myVar ; i += 1 )); do indx = " $indx 1" ; done echo $indx > index.txt Eddy
fourth dimension eddy_cuda8.0 \ --imain=dwi_xc.nii.gz \ --acqp=acqparams.txt \ --index=index.txt \ --mask=dwi_bse_mask.nii.gz \ --bvecs=dwi_xc.bvec \ --bvals=dwi_xc.bval \ --out=dwi_eddy \ --cnr_maps \ --repol \ --mporder=xvi \ --estimate_move_by_susceptibility \ --verbose One mode to check eddy correction is to create a gif image that scans through each volume forth the same slice. I've create an R script that can salve a gif for each participant in your report:
library(oro.nifti) library(neurobase) setwd('/fslhome/intj5/compute/information/BRAINSCOPE/pnlNipype/') dirs= list.files(path= ".") for (ten in ane : length(dirs)){ setwd(paste('/fslhome/intj5/compute/data/BRAINSCOPE/pnlNipype/', dirs[x], sep = "")) getwd() img <- readNIfTI("dwi_eddy.nii.gz") png(file= "eddy%02d.png", width= 200, height= 200) for (i in 1 :img@dim_[5]){ oro.nifti:: prototype(robust_window(img),z = 35,due west = i,plot.blazon = "single") } dev.off() system("catechumen -delay 8 *.png eddy.gif") file.remove(list.files(pattern= ".png")) } 
Eddy Quality Control
For full information: https://fsl.fmrib.ox.ac.united kingdom/fsl/fslwiki/eddyqc/UsersGuide
Participant
for i in $( ls <path-to-dir>/pDWI/ | grep "sub-" ) ; do SUBJ_DIR =<path-to-dir>/pDWI/${i} eddy_quad \ ${SUBJ_DIR}/dwi_eddy \ -idx ${SUBJ_DIR}/index.txt \ -par ${SUBJ_DIR}/acqparams.txt \ -chiliad ${SUBJ_DIR}/dwi_b0_mask.nii.gz \ -b ${SUBJ_DIR}/dwi.bval \ -5 done Study
eddy_squad \ participants.txt \ # Listing with full-path proper noun to dwi_eddy.qc directories -g grouping.txt \ -u \ -o squad_site The correct way to format the group.txt file:
VarName 0 [1 for continuous] v_1 v_2 . . . v_n Notation that the variable must always be numeric regardless if the variable is categorical or continuous.
EPI Baloney Correction
Create a skull stripped B0 image
It is easier to register the B0 and T1 if they are appropriately skull stripped. The skull will cause registration errors.
Fix Displacement between B0 and T1
Do a rigid alignment between preprocessed T1 and boil corrected B0 image to eliminate whatsoever displacement / motion differences betwixt the two images.
antsRegistrationSyNQuick.sh \ -d iii \ -f b0.nii.gz \ -one thousand t1.nii.gz \ # skull stripped image from FreeSurfer -o t1tob0- \ -t a Fix Distortions
Non-linearly register B0 to T1 to fix distortions (stretching and squishing). You are not moving the b0 image, only adjusting voxels out of place. You do not need to adjust bvec file afterwards.
antsRegistrationSyNQuick.sh \ -d 3 \ -f t1tob0-Warped.nii.gz \ -k b0.nii.gz \ -o epi- \ -t so Employ Transformation
Utilise the registration to the entire eddy electric current corrected paradigm.
antsApplyTransforms \ --dimensionality 3 \ --input-image-type 3 \ --input eddy.nii.gz \ --output epi.nii.gz \ --reference-image t1tob0-Warped.nii.gz \ --interpolation Bspline \ --transform epi-1Warp.nii.gz Topup
Create a Symlink for all your files:
ln -fs ${BIDS_DIR}/dwi/${i}_${2}_acq-AP_dwi.nii.gz AP_scan.nii.gz ln -fs ${BIDS_DIR}/dwi/${1}_${2}_acq-PA_dwi.nii.gz PA_scan.nii.gz ln -fs ${BIDS_DIR}/dwi/${1}_${2}_acq-AP_dwi.bval AP_scan.bval ln -fs ${BIDS_DIR}/dwi/${1}_${2}_acq-AP_dwi.bvec AP_scan.bvec ln -fs ${BIDS_DIR}/dwi/${1}_${ii}_acq-PA_dwi.bval PA_scan.bval ln -fs ${BIDS_DIR}/dwi/${1}_${2}_acq-PA_dwi.bvec PA_scan.bvec Extract B0 for each scan and combine together:
fslroi AP_scan b0_AP_scan 0 1 fslroi PA_scan b0_PA_scan 0 1 fslmerge -t b0_AP_PA_scan b0_AP_scan b0_PA_scan Create an acqparams.txt file:
printf "0 -i 0 0.0805839" > acqparams.txt printf "\n0 1 0 0.0805839" >> acqparams.txt Run topup:
topup \ --imain=b0_AP_PA_scan \ --datain=acqparams.txt \ --out=topup \ --config=b02b0.cnf \ --iout=b0_unwarped Create a brain mask:
fslmaths b0_unwarped -Tmean b0_unwarped_mean bet b0_unwarped_mean b0_unwarped_brain -yard -f 0.1 Create a single 4D image:
fslmerge -a AP_PA_scan AP_scan PA_scan Combine the bvec and bval files:
paste -d " " AP_scan.bvec PA_scan.bvec >> bvecs paste -d " " AP_scan.bval PA_scan.bval >> bvals Create index.txt:
myVar = ($( wc -west AP_scan.bval)) indx = "" for (( i = 1 ; i <= $myVar ; i += 1 )); do indx = " $indx 1" ; done for (( i = 1 ; i <= $myVar ; i += 1 )); exercise indx = " $indx ii" ; done echo $indx > index.txt Run eddy:
eddy_openmp \ -v \ --imain=AP_PA_scan \ --mask=b0_unwarped_brain_mask \ --index=alphabetize.txt \ --acqp=acqparams.txt \ --bvecs=bvecs \ --bvals=bvals \ --topup=topup \ --out=eddy \ --cnr_maps \ --repol \ --mporder=sixteen \ --estimate_move_by_susceptibility \ At this point you do not demand to run any epi distortion corrections since topup was included with eddy to set up the distortions.
Generate New Mask
A concluding mask needs to be generated earlier tensor fitting.
How To Register A 4d Image Fsl Diffusion,
Source: https://bookdown.org/u0243256/tbicc/preprocessing-diffusion-images.html
Posted by: williamsbour1950.blogspot.com


0 Response to "How To Register A 4d Image Fsl Diffusion"
Post a Comment